Description
Zebrafish (Danio rerio) are an attractive model organism for the study of human disease pathology because of their optical transparency and genetic tractability. They serve as a great alternative to mammalian screening due to cost, throughput and reduced ethical concerns.
Automated analysis of Zebrafish imposes unique demands due to the versatility of organs and features needed to be detected.
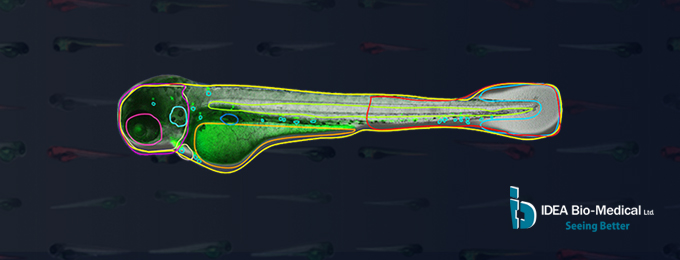
To increase throughput, robustness and permit unbiased image analysis of zebrafish, IDEA Bio-Medical developed a novel AI-based software to automatically detect fish contour and internal anatomy in brightfield images. By identifying these hard to detect structures, our software maintains the anatomical context of associated fluorescence signals to enable true high-content imaging in Zebrafish. Thusly, zebrafish morphology along with region-specific spot/cell counting and fluorescence intensity measurement are readily quantified.
· Quick & simple user interface
· Designed for non-specialists in image analysis
· Compatible for images from commercial microscopes
· The fastest & easiest Zebrafish image-based screening tool available
· Novel Artificial Intelligence- based algorithms for fish & organ identification
· No user intervention required to readily detect fish & internal organs in brightfield
· Combine with fluorescent labeling for structural co-analysis
· Ensure proper fish orientation in post-analysis with customizable, software-based selection
· Software suited for researchers who only image several fish per week, as well as researchers imaging hundreds & thousands of fish in multi-well plates for large scale screens.
· Software accepts any microscopy image, supporting multiple image formats from manual microscopes or automated imaging systems from vendors such as Leica, Zeiss, Nikon and more.
· Unbeatable throughput: analyze hundreds of Zebrafish larvae within minutes
· Analyzes Label-free or fluorescently tagged fish and internal organelles
· Analyze your Fish larva from head to tail with supported image analysis from single plane, Z stack and projection images
Technical Specifications:
Computer requirements
- Hard Disk space available: 6GB minimum.
- Minimum 8GB RAM, preferable 16GB.
- At least 2 cores, preferable 8.
- Monitor screen: To ensure proper software visibility, screen resolution should be set to at least 1920×1080 and the text scaling should be set to 100%.
Image formats
Athena Zebrafish supports loading a broad range of image formats either with or without the associated Metadata. Images must be either 8-bit or 16-bit grayscale images, as RGB color images are not supported.
Without metadata, supported image formats are *.tif, *.tiff, *.png, *.bmp, *.jpg, *.jpeg. For these formats, users can manually define pixel size and the software can identify associated color channels present in separate image files. To group the color channels together, identifying text in the file name of each image should be present e.g., Brightfield_01.tif, Green_01.tif, Red_01.tif. Multi-page *.tif image stacks are accepted when each page corresponds either to a Z- or T-stack (time-lapse). Each Z- or T-slice is analyzed separately and 3D rendering is not supported.
When loading OME-TIFF and proprietary formats (*.czi, *.nd2, etc), Athena can automatically extract the following associated metadata:
- Dimensions of each image: well, in well field, wavelength, time index, Z-stack index.
- Pixel size
- Plate info
- Objective
Image Characteristics
Athena Zebrafish has four requirements to visualize zebrafish embryos or larvae visualized in a micrograph image:
- Analysis of Zebrafish larvae and embryos is supported, up to 5 DPF (5 days post fertilization).
- The zebrafish animal must be imaged using transmitted light for contour and anatomy detection. Whole-body fluorescence images are not supported.
- The transmitted light channel must have a bright (white) background. Best results are obtained for zebrafish imaged with standard brightfield illumination rather than contrast enhanced transmitted light methodologies (i.e. phase contrast, DIC).
- The whole animal must be visible within the image, from head to tail. Images visualizing only specific regions, such as the heart or tailfin, are not yet supported.